Hi,
I am trying to compute NOs and NTOs from a RAS-2SF calculation for water using the following input:
$molecule
0 5
O -2.1904701 0.5202006 0.0237374
H -1.2011437 0.4799213 0.0090967
H -2.4798356 -0.3698583 -0.2997841
$end
$rem
JOBTYPE = SP
EXCHANGE = HF
CORRELATION = RASCI
BASIS = 6-31G
UNRESTRICTED = FALSE
SCF_ALGORITHM = DIIS
SCF_CONVERGENCE = 8
THRESH = 14
RAS_ACT = 4
RAS_ELEC = 4
RAS_OCC = 3
RAS_ROOTS = 5
RAS_SPIN_MULT = 0
RAS_PRINT = 3
RAS_AMPL_PRINT = 10
SET_ITER = 500
GUI 2
state_analysis 1
nto_pairs 3
$end
but the .fchk file doesn’t seem to contain either of the 2 types of orbitals when using IQmol.
Using MOLDEN_FORMAT = 1 I can save NTOs but still no NOs.
I tried using the RAS_NATORB_STATE = 1 (or higher) but I still cannot plot NOs.
Can someone point out what I am missing or a more convenient way to compute all orbitals? Are cube files implemented?
Hi Goran, I used “RAS_NATORB_STATE = true” with your $rem and it does save NOs (with NTOs) in the .fchk. So your question pertains to plotting them with IQmol, correct?
Hi Kaushik,
maybe the issue is with plotting only. According to the manual, RAS_NATORB_STATE takes an integer that specifies for which state you want NOs, but if using it with a boolean returns NOs for all states then I will use it as such.
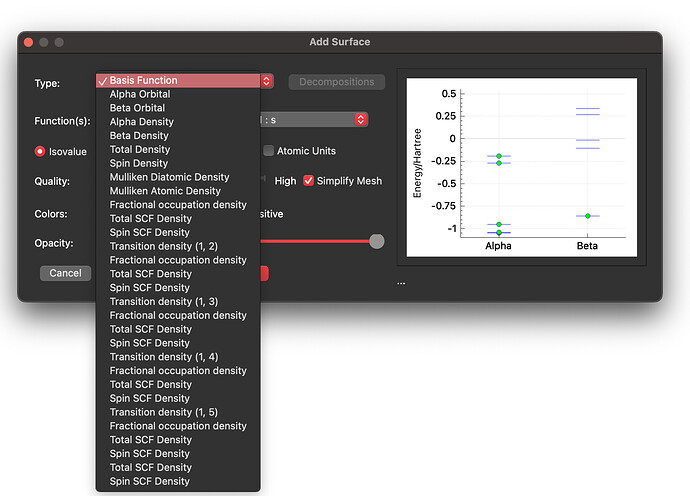
Anyway, in IQmol I do not see NOs or NTOs in the list of available surfaces, see below:
Do you plot them in a different way?
There is a workaround for NTOs. If you comment out GUI = 2, the NTOs are copied in the <working-directory>/<output-name>.plots folder. You may use these _nto.mo files for visualizing NTOs (similar to how we get NTOs for coupled-cluster methods). NOs in _no.mo files are not being generated though.
Yes, indeed that’s what I mention in my first post. I can get NTOs in molden format but not the NOs. The thing is, I would like to see the NOs for the reference and excited states to see where the unpaired electrons are located.
I guess we could interface that to libwfa at some point. And then we have everything of interest together
Goran, there might still be a workaround for NOs, which you can test if it is absolutely urgent. Please run a GUI=2 calculation with TDDFT with singlet or triplet water with STATE_ANALYSIS = 1 and NTO_PAIRS = 4. In the TDDFT .fchk file, note the header strings (“Alpha NTO coefficients …” and “Alpha NTO amplitudes …”) before the printout of NTOs singular values and coefficients for each transition. These header strings are likely parsed in IQmol and are different than those printed in RAS .fchk file currently. If you load the TDDFT .fchk in IQmol, it does recognize the NTOs and is able to plot them. So, if it is pressing, you can use the format of the TDDFT NTO header strings and replace the header strings for NOs in the RAS .fchk file manually and load them in IQmol. Note that there might be more rearrangements to be done since both the singular values and coefficients for hole and particle NTOs are grouped together, whereas in the RAS .fchk file, hole and particle NTO information is printed separately. Hope that makes sense.
We have noted this unfortunate issue with RAS .fchk file and will resolve it in future versions of Q-Chem and IQmol. Thank you for pointing it out.
Kaushik, it’s funny you mentioned this, I had the same idea this morning but I changed the headers for Alpha and Beta MOs since there was a better similarity in format:
MOs coefficient → NOs coefficient
MOs energy → NOs occupation
Not ideal, but I think it works for now. Water was just an example, but I’ll see how it goes for my system.
Thank you all for the discussion