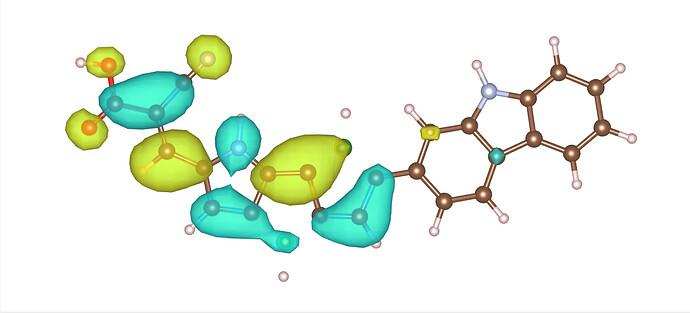
Hello,
I was trying to plot the orbitals with the following input file, but after obtaining the cube file and drawing the MO orbital, I found that some of the bonds are broken. Is there something that I forgot to mention in the input? I have attached an image of the orbital for your reference
$comment
B_B_N
$end
$molecule
0 1
C -3.64564038609275 1.83337422660860 -0.17604433089520
C -2.68373973024537 0.76502587561901 -0.08071554277843
C -3.16023314834343 -0.57790299712809 0.02488634358201
C -4.53361653412957 -0.80534917977629 0.03176325529673
C -5.48966917904975 0.26583541096779 -0.06447143336131
C -5.01603157686745 1.59525313684468 -0.16879966056003
H -3.28634457465272 2.86711186977596 -0.25771112717236
H -2.43317695148276 -1.39809576145054 0.09789241701237
H -5.72503850693656 2.43270927575366 -0.24356149374084
H -4.80582573488814 -2.92674760882751 0.20099880020613
C -6.59779887061757 -1.75615087912100 0.08999551193522
C -7.67741141486512 -2.65517766514389 0.15257479524255
C -8.97038722345173 -2.11762320759455 0.09541618162776
C -9.19175707184901 -0.72170366965776 -0.02109043803627
C -8.11736249966471 0.17085734934416 -0.08303411880798
C -6.80030598774573 -0.33813166939460 -0.02776310194784
H -7.51627618171116 -3.73929438708282 0.24251128891216
H -9.83400529701039 -2.79733614543773 0.14206266157428
H -10.22232976732582 -0.34121907318079 -0.06288943002485
H -8.29188552674957 1.25294975589719 -0.17382223843594
N -5.23360041504062 -2.00585157089556 0.12360045668709
C 3.10047176043031 2.20325670371706 -0.15468422536423
C 2.52364981882047 0.85049773201559 -0.05545132652628
C 1.13050448276211 0.80763166435571 -0.05817049725198
C 0.70091093738016 2.30672455315759 -0.17419400602009
C -1.25882509476805 1.00047251181443 -0.08816876302311
C -0.67165827412751 2.35246609793045 -0.18724670282928
H -1.25160585746819 3.28457743474544 -0.26201989950598
C 4.49643661331338 2.02348067459939 -0.13646472803583
C 4.77038669855897 0.62945808699393 -0.03154179026457
H 5.27928344022497 2.78940278317069 -0.19139505256100
C 6.06792397557742 0.04275798418670 0.01548355792613
C 6.42668843888871 -1.29295200838533 0.11403050478400
H 6.91767247754140 0.74170332935689 -0.03363851790093
C 5.46692122429981 -2.34441265665032 0.18897485807046
C 7.88267151149102 -1.61972849127142 0.14056845138750
N 4.62760185148683 -3.17382671217022 0.24783543966197
O 8.78164571896789 -0.79011291535028 0.08287378958553
O 8.09853730145440 -2.96118561306295 0.23721764381417
H 9.07810083565297 -3.06259811102473 0.24662227497979
H 3.46511579634466 -1.07556269246955 0.09097957384610
H -0.20623314980798 -1.25717324464769 0.09106544882713
N 3.55533133082771 -0.05835816412522 0.01613218447693
B 1.93939926538877 3.22919192752083 -0.23696836028783
H 1.99573761820653 4.43428092696657 -0.32702848252956
B -0.10319836172672 -0.05229974949350 0.00006374742575
$end
$rem
SCF_CONVERGENCE 6
MAX_SCF_CYCLES 100
MAKE_CUBE_FILES true
PLOTS true
EXCHANGE gen
LRC_DFT = true
omega = 219
CIS_N_ROOTS 1
CIS_SINGLETS true
CIS_TRIPLETS true
RPA FALSE
BASIS def2-TZVPD
STATE_ANALYSIS true
MEM_TOTAL 2000000
$end
$xc_functional
C PBE 1.00
X wPBE 1.00
$end
$plots
grid_points 50 50 50
alpha_molecular_orbital 100-107
$end