I am using iqmol interface to perform qchem calculations on protonated and deprotonated histidine. I am a bit concerned because I create my molecules in chemdraw then import via SMILES code. After I run the qchem calculation my output structure seems incorrect, making me question the electrostatic protential surface (which is what im interested in). Shouldn’t there be a double bond between N6 and C7?
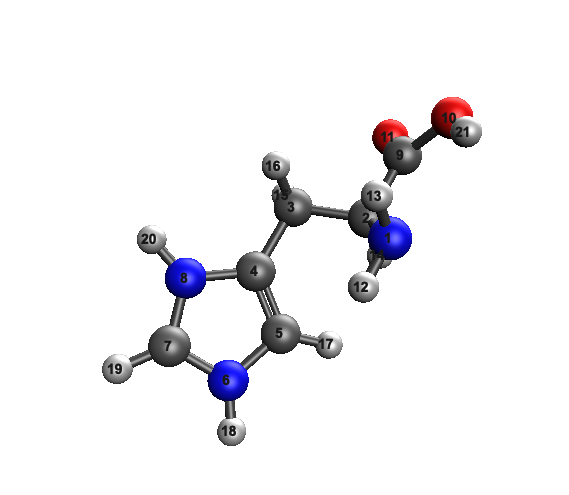
Hi there, did you infer that your structure is incorrect simply based on whether N6–C7 bond is displayed as a double bond in IQmol? Each software has its own definition of a double bond. IQmol not displaying a double bond between N6 and C7 does not necessarily mean there isn’t one.
Ah got it. I will look into the iqmol features. I guess my conventional knowledge tells me there should still be a double bond there despite the charge.
Thanks!
You can draw a double bond if you want, it’s just lines on a page without the electronic structure.
1 Like