Hello,
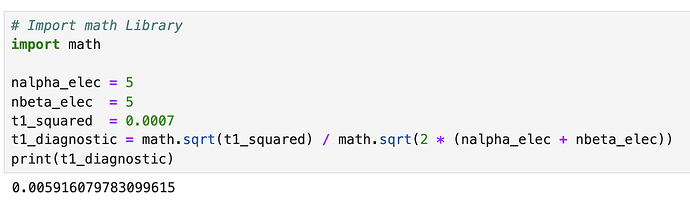
After running CCSD(T) calculation for H2O molecule, I was looking into the output file to find T1 diagnostic value. Can anyone show me where to look? There is only T1^2 value after reporting energies and I don’t know what is referring to.
Thank you for your time.
Input file:
$molecule
0 1
H -2.3527135 2.2827770 0.4401250
O -3.1357501 2.2827770 -0.1660574
H -3.9187867 2.2827770 0.4401250
$end
$rem
jobtype = sp
method = ccsd(t)
basis = def2-SVP
n_frozen_core = fc
ccman2 = true
$end
Output file:
Starting (T) calculation…
Using double precision libpt code
Running restricted (T) code
Using 1 integral batch
(T) calculation completed in 0.01 sec
(T) energy is -0.0031326630
SCF energy = -75.95721338
MP2 energy = -76.16060275
CCSD correlation energy = -0.21302039
CCSD total energy = -76.17023378
CCSD(T) correlation energy = -0.00313266
CCSD(T) total energy = -76.17336644
CCSD T1^2 = 0.0007 T2^2 = 0.0588 Leading amplitudes:
Amplitude Orbitals with energies
0.0097 3 (A1) A → 4 (A1) A
-0.5621 0.1710
0.0097 3 (A1) B → 4 (A1) B
-0.5621 0.1710
0.0075 1 (B1) A → 6 (B1) A
-0.6863 2.4830
0.0075 1 (B1) B → 6 (B1) B
-0.6863 2.4830
Amplitude Orbitals with energies
-0.0512 1 (B2) A 1 (B2) B → 2 (B2) A 2 (B2) B
-0.4957 -0.4957 1.2016 1.2016
0.0512 1 (B2) A 1 (B2) B → 2 (B2) B 2 (B2) A
-0.4957 -0.4957 1.2016 1.2016
0.0512 1 (B2) B 1 (B2) A → 2 (B2) A 2 (B2) B
-0.4957 -0.4957 1.2016 1.2016
-0.0512 1 (B2) B 1 (B2) A → 2 (B2) B 2 (B2) A
-0.4957 -0.4957 1.2016 1.2016